- *Corresponding Author:
- M. Salihovic
Department of Pharmacy, University of Sarajevo, Sarajevo 71000, Bosnia and Herzegovina
E-mail: mirsada.salihovic@ffsa.unsa.ba
| Date of Received | 16 February 2022 |
| Date of Revision | 16 March 2023 |
| Date of Acceptance | 21 June 2023 |
| Indian J Pharm Sci 2023;85(3):769-777 |
This is an open access article distributed under the terms of the Creative Commons Attribution-NonCommercial-ShareAlike 3.0 License, which allows others to remix, tweak, and build upon the work non-commercially, as long as the author is credited and the new creations are licensed under the identical terms
Abstract
In the reaction of 3-aminothymoquinone and aromatic aldehydes, two benzoxazole derivatives viz. 2-(4-methoxyphenyl)-4-methyl-7-isopropyl-1,3-benzoxazol-5-ol (1) and 2-(4-trifluoromethyl)-4-methyl-7-isopropyl-1,3-benzoxazol-5-ol (2) were prepared and characterized by elemental analysis, infrared and 1H, and 13C-nuclear magnetic resonance spectroscopy and mass spectrometry. Their antimicrobial activity against Escherichia coli, Salmonella enterica, Proteus hauseri, Pseudomonas aeruginosa, Staphylococcus aureus, Sarcina lutea, Clostridium sporogenes and Bacillus subtilis was tested. Synthesized compounds show the best activity on Sarcina lutea, and the lowest against Proteus hauseri and Clostridium sporogenes. The paper assesses in silico methods of the possible ways selected derivatives bind to the enzyme deoxyribonucleic acid gyrase (1KZN). The docking results were compared with those obtained from in vitro antimicrobial activity. Molecular properties and absorption, distribution, metabolism and excretion parameters were also calculated for compounds. The difference in the obtained values reflects differences in the derivatives structures. In the future, tests on a number of enzymes crucial for bacterial life as well as a number of derivatives may offer further information on the mechanisms of action of these substances.
Keywords
Benzoxazoles, antimicrobial activity, docking, molecular properties
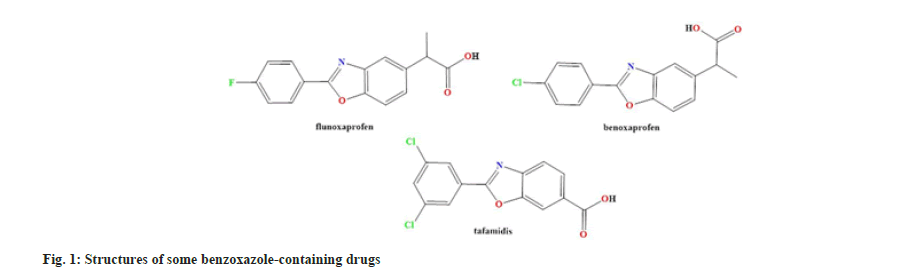
Benzoxazoles are molecules with a wide range of biological activities, including antimicrobial[1], antitumor and anti-inflammatory activities[2]. For obtaining benzoxazoles, numerous synthetic routes have been reported in the literature; Cyclization of Alpha (α)-Oxo Oxime[1], then condensation of o-aminophenol with carboxylic acids, oxidative cyclization of phenolic Schiff bases[3], and condensation reactions of o-aminophenol with orthoesters, o-haloamides and 1,1-dihaloalkenes. Particular attention has been directed to finding the most suitable catalysts in terms of cost, availability and environmental friendliness[2-4]. Benzoxazole is an integral part of many drugs on the market as shown in fig. 1[5].
Synthesizing benzoxazole derivatives and testing their antimicrobial activity has become a very interesting area for scientists, especially in recent studies[6]. On the other hand, inhibition of bacterial enzymes is a well-known mechanism by which drugs induce antibacterial activity. Deoxyribonucleic Acid (DNA) gyrase (topoisomerase II) is an essential bacterial enzyme that catalyzes the Adenosine Triphosphate (ATP) dependent negative super-coiling of double-stranded closed-circular DNA. Much attention has been focused on DNA gyrase as an intracellular target of several antibacterial agents[7]. It is an essential enzyme found in all bacteria but lacking in higher eukaryotes, making it an attractive antibacterial target[8].
Several in silico methods can be used to shed light on potential targets and interactions of novel small molecules with defined target macromolecules (receptor). Molecular docking is a popular method to study the binding of interest[9,10].
So far, for compounds of this type, docking has been performed on target enzymes important for the growth and development of tumor cells[11], and no enzymes essential for bacterial life. Except docking, numerous other parameters, that provide information about the potency of some compounds to become drugs, were investigated in silico.
In view of the foregoing, the synthesis, structural characterisation and antibacterial activity of two Benzoxazoles derivatives obtained from aromatic aldehydes with 3-Aminothymoquinone (ATQ) are presented in this study. Docking studies of both synthesized compounds were analyzed based on the in vitroantibacterial results in order to evaluate the correlation between in silico studies and in vitro antimicrobial results.
Materials and Methods
Chemistry:
Apart from ATQ, all solvents and reagents were sourced from Merck KGaA, Darmstadt, Germany. To observe the flow of reactions and the purity of the end products, aluminum or glass plates (10×10 cm) coated with silica gel 60 F254 (Merck KGaA, Darmstadt, Germany) were used. The mobile phase was selected depending on the mixture being tested. For detection, Ultra Violet (UV) light of 254 nm was used. Synthesized compounds were purified by column chromatography on Silica gel 60 (Merck KGaA, Darmstadt, Germany). The choice of the optimal mobile phase was made according to the solvent selection guidelines by Talamona et al.[12]. For the substance to be purified, the aim was to obtain an Retardation Factor (Rf) value of 0.15-0.4 at Thin Layer Chromatography (TLC) analysis.
Melting points were determined on a Kofler Hot Stage Microscopes Polyether apparatus, Meerhout, Belgium and are uncorrected. The elemental analysis was accomplished by combustion analysis on a Vario EL III C, H, N, S/O Elemental Analyzer, Germany (Elementar). The mass spectra were recorded with an Agilent 6420 Triple Quadrupole mass spectrometer with an electrospray ionization source, Santa Clara, United States. The Fourier Transform Infrared (FTIR) spectra were obtained in the range 4000-450 cm-1 on a FTIR attenuated total reflection PerkinElmer UATR two spectrometer, Ohio, United States.
Nuclear Magnetic Resonance (NMR) spectra in solution were measured in Dimethyl sulfoxide-d6 (DMSO-d6) on a Bruker AV300 spectrometer (United Kingdom) at 298 K in 5 mm NMR tubes. 1H NMR spectra were acquired at 300.131 MHz, while 13C Attached Proton Test spectra were acquired at 75.475 MHz. Digital resolutions in 1H and 13C spectra were 0.37 and 0.60 Hz per point. Chemical shifts (δ/ppm) in 1H spectra were referred to the methyl protons of Tetramethylsilane (TMS); δ=0.0 ppm. Chemical shifts (δ/ppm) in 13C spectra were referred to the signal of DMSO-d6; δ=39.51 ppm; XYZ® tablets.
Procedure for the preparation of compounds (1) and (2):
To produce ATQ, thymoquinone (1 mmol) and sodium azide (1.3 mmol) were mixed in absolute ethanol. As a catalyst, glacial acetic acid was used. Afterwards when, the mixture was refluxed at 80° for 3 h. TLC was used to monitor the progress of the reaction. The reaction was stopped by adding Sodium bicarbonate (NaHCO3) to the mix. The crude reaction mixture was purified using column chromatography with silica gel 60 as the stationary phase and Dichloromethane (DCM):Ethanol:Hexane=1:2:5 (v/v/v) as the eluent[11,13].
In absolute ethanol, 4-methoxybenzaldehyde or 4-(trifluoromethyl) benzaldehyde was added in equimolar amounts to synthesize ATQ with HCl as a catalyst. The mixture was then refluxed for 4 h at 80° to produce compound (1) or (2). The mixture was poured over crushed ice, filtered, washed with distilled water, and purified utilizing column chromatography with silica gel 60. Purification of compounds necessitated the use of a different¢s mobile phase.
The mobile phase DCM:EA was used to extract pure ATQ on a column at a ratio 20:0.3 (v/v)[11]. This mobile phase did not prove to be good for the separation of compound (2). DCM:Hex:EA in a ratio of 2:5:1 (v/v/v) is a system that has shown good separation. System DCM:Hex:EA in a ratio of 2:5:0.3 (v/v/v) was even more suitable because the stains are better separated, which is conducive to column purification. The solvent system described in our previous studies was used to purify compound (1)[11].
7-isopropyl-2-(4-methoxyphenyl)-4-methylbenzo[d]oxazol-5-ol (1)[11]: White solid (0.130 g, 43.7 %); m.p. 178 C; 1H-NMR (DMSO-d6) 9.11 (1H, s, OH), 8.11 (2H, d, JH,H=7.98 Hz, H-12, H-16), 7.14 (2H, d, JH,H=7.98 Hz, H-13, H-15), 6.71 (1H, s, H-6), 3.86 (3H, s, Ethyl Methyl ether (OCH3)), 3.24 (1H, hept, JH,H=6.65 Hz, H-8), 2.32 (3H, s, CH3-17), 1.34 (6H, d, JH,H=6.95 Hz, CH3-9, CH3-10); 13C-NMR (DMSO-d6) 161.74 (C, C-14), 161.67 (C, C-2), 152.1 (C, C-5), 141.8 (C, C-3), 141.4 (C, C-1), 128.7 (CH, C-12, C-16), 127.9 (C, C-7), 119.4 (C, C-11), 114.6 (CH, C-13, C-15), 111.3 (C, C-4), 110.0 (CH, C-6), 55.4 (CH3, C-19), 28.8 (CH, C-8), 22.5 (CH3, C-9, C-10), 10.1 (CH3, C-17); HRESIMS m/z 298.14311 [M+H]+ Anal. Calcd. for C18H19NO3: C 72.71, H 6.44, N 4.71 %. Found: C 71.96, H 6.43, N 4.62 %.
2-(4-trifluoromethylphenyl)-4-methyl-7-isopropyl-1,3-benzoxazol-5-ol (2): Yellow solid (0.090 g, 30 %); m.p 189.3°; IR (ATR) νmax/cm–1: 2968, 1503, 1325, 1166, 751; 1H-NMR (DMSO-d6) δ/ppm: 9.27 (1H, s, OH), 8.37 (2H, d, J=8.22 Hz, H-12 and H-16), 7.96 (2H, d, J=8.22 Hz, H-13 and H-15), 6.81 (1H, s, H-6), 3.28 (1H, hept, J=6.90 Hz, H-8), 2.36 (3H, s, CH3-17), 1.36 (6H, d, J=6.96 Hz, CH3-9 and CH3-10); 13C NMR (DMSO-d6) δ/ppm: 160.1 (C-2), 152.4 (C-5), 141.9 (C-3), 141.6 (C-1), 130.6 (q, JC,F=32.0 Hz, C-14), 130.4(d, JC,F=1.2 Hz, C-11), 128.4 (C-7), 127.7 (C-12 and C-16), 126.2 (q, JC,F=3.6 Hz, C- 13 and C-15), 123.9 (q, JC,F=272.0 Hz, CF3), 111.9 (C-4), 111.4 (C-6), 28.8 (C-2), 22.4 (C-9 and C-10), 10.1 (C-17). ESI (+)-MS m/z: 336.9 [C18H16F3NO2+ H]+. Anal. Calcd. for C18H16F3NO2: C 64.47; H 4.81; N 4.18 %. Found: C 64.36; H 4.70; N 4.15 %.
Antimicrobial activity of synthesized compounds:
Antimicrobial activity of synthesized compounds was performed by using eight different bacterial strains: Escherichia coli (E. coli) ATCC 25922), Salmonella enterica subsp. Enterica serovar Enteritidis (ATCC 13076), Proteus hauseri (P. hauseri) (ATCC 13315), Pseudomonas aeruginosa (ATCC 9027), Staphylococcus aureus (S. aureus) ATCC 6538), Sarcina lutea (S. lutea) (ATCC 9341), Clostridium sporogenes (C. sporogenes) (ATCC 19404) and Bacillus subtilis (ATCC 6633).
For the determination of the Minimum Inhibitory Concentration (MIC) of the synthesized compounds, the micro dilution method was used. Cultures of test strains were grown in nutrient agar. Grown cultures were then suspended in a sterile physiological solution. Obtained suspension contained 1.5×108 CFU/ml and had opacity of 0.5 McFarland standards (National Committee for Clinical Laboratory Standards (NCLS), 2003). Stock solutions of synthesized compounds having a concentration of 10 mg/ml in dimethyl sulfoxide were prepared and then diluted in a 96-well microliter plate. In the first well, the concentration of the test sample has already been reduced to half (i.e., 5 mg/ml) as the stock solution had been diluted with an equal volume of the medium. After that, 100 µl of the solution was transferred from the first well to the second and the process is repeated until the end of the line. Wells containing the medium and tested sample were inoculated with cultures of microorganisms, and the microliter plates were incubated at 37° for 24 h for bacteria. After thermosetting, in wells where the growth of the microorganisms was obtained, diffuse turbidity could be seen. On contrary, wells with no microbial growth remained clear. The illuminator determined whether there was turbidity in a well.
Docking studies:
The docking studies were performed with the aim of determination of probable antimicrobial activity mechanisms. The crystal structure of DNA gyrase (Protein Data Bank (PDB) ID: 1KZN) from PDB was used as the target molecule. The protein structure was prepared by removing water molecules, adding polar hydrogen atoms and optimizing in the AMBER03 force field. Structures of tested compounds were optimized in Chem3D Ultra software 16.0.1.4. with the use of the MM2 method. The docking study was set up in YASARA software 19.12.14[14,15] and performed using AutoDock 4.2[16]. Coordination x, y, z network with dimensions 136×154×140 dots, and distance between dots 0.375 Å was prepared for blind docking approach, searching the whole protein for potential binding sites. Lamarckian Genetic Algorithm (LGA) was used for searching conformers that are energetically the most favorable, with cluster tolerance of 5 Å. Up to 150 conformers of each compound were analyzed. The maximal number of function evaluations was 12 500 000, the maximal number of 27 000 generations, with the number of individuals restricted to 1, mutation rate 0.02, crossover rate 0.8, 2 Å translation, quaternion angle 50 degrees and torsion angle 50 degrees. External grid energy 1000, maximal initial energy 0 and the maximal number of repetitions 10 000. Visualization, graphical rendering, and structural analysis were performed using YASARA software[14,15].
Prediction of molecular and Absorption, Distribution, Metabolism and Excretion (ADME) descriptors:
Prediction of molecular, pharmacokinetic properties and toxicity is very important in the current drug development process. Log P (lipophilicity), number of Hydrogen Bond Donors (HBD), number of Hydrogen Bond Acceptors (HBA), topological polar surface area as well as the bioactivity scores, i.e., the affinity of the compounds for various targets such as G-protein coupled receptor, ion channel, kinase, nuclear receptor, protease, and enzyme were predicted using an online tool (https://www.molinspiration.com). ADME properties of the title compounds were predicted using the preADMET online server (https://preadmet.bmdrc.org/). Various ADME properties including Colorectal Carcinoma (CaCo-2) cell permeability, human intestinal absorption, plasma protein binding, blood-brain barrier penetration, skin permeability, and Maden Darby canine kidney cell permeability were predicted.
Results and Discussion
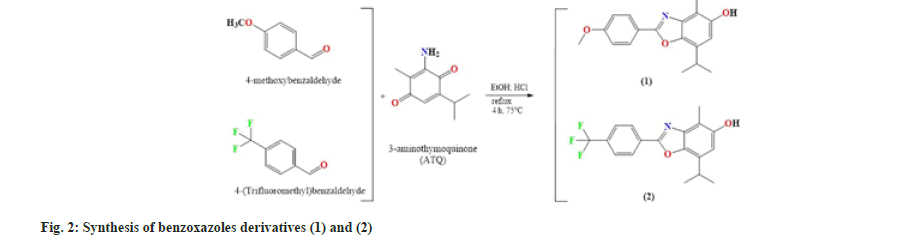
According to the processes described in fig. 2, the condensation of 4-methoxybenzaldehyde and ATQ in ethanol yields compound (1), and the condensation of 4-(trifluoromethyl) benzaldehyde and ATQ in ethanol yields compound (2). Compounds (1) and (2) were synthesized in good yield (43, 7 % and 30 %, respectively). TLC on silica gel F254 as the stationary phase and dichloromethane:hexane and Ethyl Acetate (EA) as the mobile phase (2:5:1,v/v/v) were used to monitor reaction progress and assess purity of synthesized compounds. FT-IR, 1H, and 13C-NMR spectroscopy were used to characterize the synthesized compounds (1) and (2).
Using the micro dilution method, the values of Minimal Inhibitory Concentration (MIC) were expressed in mg/ml and presented in the Table 1 and Table 2. Antimicrobial activity of synthesized compounds (1) and (2) was tested on eight bacterial strains viz, E. coli, Salmonella enterica, P. hauseri, Pseudomonas aeruginosa, S. aureus, S. lutea, C. sporogenes and Bacillus subtilis.
| Bacteria | MIC (mg/ml) | ||
|---|---|---|---|
| Compound (1) | Compound (2) | Amikacin | |
| E. coli | 0.039 | 0.01 | 0.005 |
| S. enterica | 0.313 | 0.313 | 0.008 |
| P. hauseri | 0.625 | 0.625 | 0.007 |
| P. aeruginosa | 0.313 | 0.313 | 0.05 |
| S. aureus | 0.313 | 0.039 | 0.011 |
| S. lutea | 0.02 | 0.005 | 0.002 |
| C. sporogenes | 0.625 | 0.625 | 0.015 |
| B. subtilis | 0.313 | 0.313 | 0.042 |
Table 1: MIC values (mg/ml) for synthesized compounds against tested bacteria.
| Comp. | Binding energy/kcal mol–1 | Inhibition constant/µM | Contacting receptor residues |
|---|---|---|---|
| (1) | –6.56 | 15.47 | VAL 43*, ASN 46*, ALA 47*, GLU 50*, VAL 71, GLN 72, ASP 73*, ARG 76*, GLY 77, ILE 78*, PRO 79*, ILE 90*, VAL 120*, ARG 136, THR 165*, MET 166, VAL 167* |
| (2) | –6.92 | 8.52 | VAL 43*, ASN 46*, ALA 47*, GLU50*, VAL 71, GLN 72, ASP 73, ARG76*, GLY 77, ILE 78*, PRO 79*, ILE90*, VAL 120*, ARG 136, THR165*, MET 166, VAL 167 |
Note: *Hydrophobic interaction was observed with this amino acid
Table 2: Binding characteristics of compounds (1) and (2) to DNA Gyrase (PDB ID: 1KZN) receptor, as assessed by molecular docking study.
The results were expressed as the MIC (mg/ml). The reference drug was amikacin. When it comes to compound (1), it shows the best activity against bacterial strain S. lutea (MIC 0.020 mg/ml) and very good activity towards E. coli (MIC 0.039 mg/ml). These activities are smaller, but certainly comparable to the reference drug, amikacin. This compound showed also the lowest activity against bacterial strains P. hauseri and C. sporogenes (MIC 0.625 mg/ml). A similar trend in antimicrobial activity was confirmed for compound (2). The maximum activity against bacterial strains S. lutea and E. coli, and the least one against strains P. hauseri and C. sporogenes, just as with compound (1). The reference drug has also proven to be much more effective. When comparing our two compounds, it can be concluded that compound (2) is better, because it demonstrates higher activity against bacterial strains S. lutea, E. coli and S. aureus compared to compound (1). Against S. lutea and E. coli strains, compound (2) shows about twice less activity than amikacin, while against S. aureus the activity was four times less compared to the control compound. The values of inhibitory concentrations in all other cases proved to be identical. Compound (2) possesses a lipophilic CF3 group which probably contributes to better microbiological activity.
Considering the results obtained from antibacterial activity, it was necessary to perform molecular docking studies to examine the relationship between the in vitro results and in silico studies. The comparative docking of receptor DNA gyrase, with compounds (1) and (2) exhibited good affinity. Both compounds bound to the same site within the receptor. Compound (1) showed binding energy of –6.56 kcal/mol, while compound (2) showed binding energy of –6.92 kcal/mol. There were multiple hydrophobic interactions with receptor observed for both compounds as presented in Table 2 and fig. 3.
Values of binding energies of tested compounds were similar as assessed by molecular docking study, however, somewhat lower values of binding energy and inhibition constant for compound (2) correlate in vitro results obtained where compound (2) exhibited better activity on several bacterial strains.
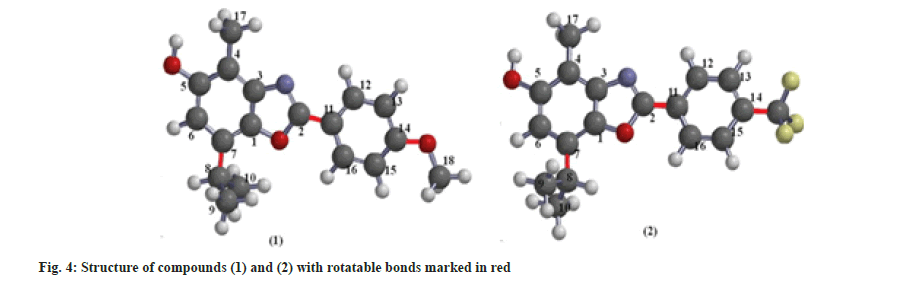
Docking does not answer the many questions that arise in the initial stages of testing for biologically active compounds. Numerous other parameters provide information about the potency of some compounds to become drugs. The ability of the drug to pass through the membrane is one of the first parameters tested in the modeling of the new drugs. Lipinski's rule of five is well known. According to Lipinski, a molecule will be able to pass the membrane by passive intestinal diffusion if it has: Molecular Weight (MW) of less than 500 g/mol a logp value of less than 5 representing its hydrophobicity, no more than 5 HBD, and no more than 10 HBA sites[17]. Further research has added two more conditions: A Polar Surface Area (PSA) of less than or equal to 140 Å and less than 10 rotatable bonds (Rotb)[18], which are correlated with drug permeability and flexibility (fig. 4).
In compliance with this set of rules, a chemical compound would act as an orally active drug-like compound on the desired target. If the molecule does not meet any two of the above requirements, it is assumed that it will not be capable of absorption by passive intestinal diffusion. From this rule, it is evident that the lipophilicity of the compounds is an important parameter in the process of intestinal drug absorption. But based on lipophilicity, not only the absorption but also the distribution, elimination and toxicity of a drug can be predicted.
The results of permeability predicting for the test compounds via passive intestinal diffusion, using Lipinski's rule of five, are shown in Table 3. All the compounds were predicted to have good oral bioavailability as their MW is <500 g mol–1, hydrogen bond donors are <5 (0), hydrogen bond acceptors are <10, number of Rotatable bonds (nRotb) are <10 and Topological Polar Surface Area (TPSA) <140 Ų.
| Comp. | TPSAa/Ų | miLogPb | MWc/g mol–1 | nONd | nOHNHe | nRotbf | MVg/ų |
|---|---|---|---|---|---|---|---|
| (1) | 55.49 | 5.01 | 297.35 | 4 | 1 | 3 | 276.93 |
| (2) | 46.26 | 5.85 | 335.32 | 3 | 1 | 3 | 282.68 |
Note: miLogP-Lipophilicity and g MV-molecular volume
Table 3: Molecular properties of tested Benzoxazoles.
TPSA is calculated as a sum of fragment contributions (O- and N- centered polar fragments are considered). Synthesized compounds have a different number of oxygen atoms (a different environment) and the same number of nitrogen atoms. Larger TPSA has compound (1). The nRotb is a measure of molecular flexibility and is important in determining the oral bioavailability of the drugs[19]. A rotatable bond is defined as any single non-ring bond, bounded to a nonterminal heavy (i.e., non-hydrogen) atom. According to some authors, it is desirable that the number of rotatable bonds ≤3, so both compounds satisfy the number of rotatable bonds[20].
Permeability through monolayers of human intestinal epithelial cells originated from human CaCo-2 and Madin−Darby Canine Kidney cells (MDCK) are widely considered to be the in vitro gold standard for assessing the uptake efficiency of chemicals into the body[21]. Permeability through MDCK cell lines is also used to estimate the effect of the Blood-Brain Barrier (BBB)[22,23]. Given that these tests are time and cost-intensive we calculated them using computer programs. Human Intestinal Absorption (HIA) is one of the important ADME properties and also one of the key steps during the drugs transporting to their targets[19]. "Poor" absorption was defined as HIA ≤30 %, "high" absorption as HIA ≥80 %, whereas "moderate" absorption was defined between these two values (30 % <HIA <79 %). The prediction of Plasma Protein Binding (PPB) is of paramount importance in the pharmacokinetics characterization of drugs, as it causes significant changes in the volume of distribution, clearance, and drug half-life. The reversible interaction between drug and plasma protein can also greatly influence the pharmacological effect of the drug because only a fraction of unbound drug can pass across cell membranes. Thus, it can be expected that drugs with high protein binding tend to have a greater half-life compared to those with lower values. The greater the drug is bound to plasma protein the less fraction of free drug is there for therapeutic effect[24]. As can be seen from Table 4, high human intestinal absorption and in vitro plasma protein binding are predicted for all the synthesized compounds.
Determination of compounds BBB permeability is a precondition for screening compounds that could take effects in the central nervous system[25]. As can be seen from Table 4, predicted in vivo blood-brain penetration for compound (1) is poor. On the other hand, compound (2), which has a CF3 group in the structure, is more than ten times larger. This is explained by the high lipophilicity of the CF3 group.
| Absorption | ||||||
|---|---|---|---|---|---|---|
| Comp. | HIAa/% | CaCo2b/nm sec-1 | PPBc/% | BBBd/cbrain cblood–1 | SKINe/logKp | MDCKf/nm sec-1 |
| (1) | 95.76 | 47.63 | 100 | 0.52 | -2.98 | 0.31 |
| (2) | 95.87 | 27.77 | 100 | 6.12 | -1.93 | 0.04 |
Table 4: ADME parameters of tested Benzoxazoles.
Skin Permeability (SKIN) is widely recognized as an essential parameter to be considered for the delivery of active substances. The permeability of many different compounds has been measured through several in vitro and in vivo techniques. Moreover, many different in silico approaches have been used to identify the correlation between the structure of the permeates and their permeability, to reproduce the skin behavior, and to predict the ability of specific chemicals to permeate this barrier[21]. Predicted skin permeability for two compounds was poor.
Two benzoxazole derivatives were obtained by the reaction of aromatic aldehydes with ATQ. The antimicrobial activity of the analyzed compounds tested by the micro dilution method showed that compound (2) (containing trifluoromethylphenyl group at position 2 of the benzoxazole ring) has greater activity against bacteria E. coli, S. aureus, and S. lutea compared to compound (1) (containing 4-methoxyphenyl group at the position 2 of the benzoxazole ring). Compound (2) shows better antimicrobial activity against bacteria and also better binding affinity (lower binding energy) to DNA gyrase. This can be explained by differences in the lipophilicity of these two compounds (compound (2) with a trifluoromethyl group is significantly more lipophilic), but also by the fact that the antimicrobial activity of a compound is the result of simultaneous action on several bacterial enzymes.
The activity of these compounds against DNA gyrase should not be crucial or at least not the most important in the mechanisms of action of these compounds against bacteria. It can be assumed that lipophilicity also plays an important role in explaining the better activity of compound (2).
High human intestinal absorption and in vitro plasma protein binding are predicted for all the synthesized compounds. With the introduction of the CF3 group on the benzene nucleus, blood-brain barrier permeability is increased significantly. Predicted skin permeability for two compounds was poor.
Tests on a number of enzymes important for bacterial life and a number of derivatives could provide additional insight into the mechanisms of action of these compounds in the future.
Acknowledgements:
The authors gratefully acknowledge the contribution of Dr. Vjekoslav Štrukil for recording IR spectra. The authors would like to thank the Ministry of Education, Science and Technological Development of Republic of Serbia (Grant No: 451-03-68/2021-14/200026) and the Federal Ministry of Education and Science of Bosnia and Herzegovina (Grant No: 05-39-3629-1/14) for financial support.
Conflict of interests:
The authors declared no conflict of interests.
References
- Anusha P, Rao JV, Mohan GK. A review on diverse biological activities of benzoxazole molecule. World J Pharm Pharm Sci 2017;6(7):1779-94.
- Chikhale RV, Pant AM, Menghani SS, Wadibhasme PG, Khedekar PB. Facile and efficient synthesis of benzoxazole derivatives using novel catalytic activity of PEG-SO3H. Arabian J Chem 2017;10(5):715-25.
- Patil MR, Bhanushali JT, Nagaraja BM, Keri RS. TiO2ZrO2 composite: Synthesis, characterization and application as a facile, expeditious and recyclable catalyst for the synthesis of 2-aryl substituted benzoxazole derivatives. Comptes Rendus Chim 2018;21(3-4):399-407.
- Patil MA, Ubale PA, Karhale SS, Helavi VB. Lemon juice: An environmentally benign catalyst for synthesis of benzothiazoles and benzoxazole derivatives in aqueous medium. Der Chem Sinica 2017;8(1):198-205.
- Rajasekhar S, Maiti B, Chanda K. A decade update on benzoxazoles, a privileged scaffold in synthetic organic chemistry. Synlett 2017;28(5):521-41.
- Ertan-Bolelli T, Yildiz I, Ozgen-Ozgacar S. Synthesis, molecular docking and antimicrobial evaluation of novel benzoxazole derivatives. Med Chem Res 2016;25:553-67.
- Reece RJ, Maxwell A. DNA gyrase: Structure and function. Crit Rev Biochem Mol Biol 1991;26(3-4):335-75.
[Crossref] [Google Scholar] [PubMed]
- Collin F, Karkare S, Maxwell A. Exploiting bacterial DNA gyrase as a drug target: Current state and perspectives. Appl Microbiol Biotechnol 2011;92(3):479-97.
[Crossref] [Google Scholar] [PubMed]
- Ehmann DE, Lahiri SD. Novel compounds targeting bacterial DNA topoisomerase/DNA gyrase. Curr Opin Pharmacol 2014;18:76-83.
[Crossref] [Google Scholar] [PubMed]
- Bradbury BJ, Pucci MJ. Recent advances in bacterial topoisomerase inhibitors. Curr Opin Pharmacol 2008;8(5):574-81.
[Crossref] [Google Scholar] [PubMed]
- Glamočlija U, Padhye S, Špirtović-Halilović S, Osmanović A, Veljović E, Roca S, et al. Synthesis, biological evaluation and docking studies of benzoxazoles derived from thymoquinone. Molecules 2018;23(12):3297.
[Crossref] [Google Scholar] [PubMed]
- Talamona A. Laboratory chromatography guide. 1st ed. Büchi Labortechnik AG: Switzerland; 2005.
- Yusufi M, Banerjee S, Mohammad M, Khatal S, Swamy KV, Khan EM, et al. Synthesis, characterization and anti-tumor activity of novel thymoquinone analogs against pancreatic cancer. Bioorgan Med Chem Lett 2013;23(10):3101-4.
[Crossref] [Google Scholar] [PubMed]
- Krieger E, Vriend G. YASARA View—molecular graphics for all devices—from smartphones to workstations. Bioinformatics 2014;30(20):2981-2.
[Crossref] [Google Scholar] [PubMed]
- Krieger E, Vriend G. New ways to boost molecular dynamics simulations. J Comput Chem 2015;36(13):996-1007.
[Crossref] [Google Scholar] [PubMed]
- Morris GM, Huey R, Lindstrom W, Sanner MF, Belew RK, Goodsell DS, et al. AutoDock4 and AutoDockTools4: Automated docking with selective receptor flexibility. J Comput Chem 2009;30(16):2785-91.
[Crossref] [Google Scholar] [PubMed]
- Lipinski CA, Lombardo F, Dominy BW, Feeney PJ. Experimental and computational approaches to estimate solubility and permeability in drug discovery and development settings. Adv Drug Deliv Rev 2012;64(1-3):4-17.
[Crossref] [Google Scholar] [PubMed]
- Veber DF, Johnson SR, Cheng HY, Smith BR, Ward KW, Kopple KD. Molecular properties that influence the oral bioavailability of drug candidates. J Med Chem 2002;45(12):2615-23.
[Crossref] [Google Scholar] [PubMed]
- Yan A, Wang Z, Cai Z. Prediction of human intestinal absorption by GA feature selection and support vector machine regression. Int J Mol Sci 2008;9(10):1961-76.
[Crossref] [Google Scholar] [PubMed]
- Congreve M, Carr R, Murray C, Jhoti H. A rule of three for fragment-based lead discovery? Drug Discov Today 2003;8(19):876-7.
[Crossref] [Google Scholar] [PubMed]
- Artursson P, Palm K, Luthman K. Caco-2 monolayers in experimental and theoretical predictions of drug transport. Adv Drug Deliv Rev 2001;46(1-3):27-43.
[Crossref] [Google Scholar] [PubMed]
- Summerfield SG, Read K, Begley DJ, Obradovic T, Hidalgo IJ, Coggon S, et al. Central nervous system drug disposition: The relationship between in situ brain permeability and brain free fraction. J Pharmacol Exp Ther 2007;322(1):205-13.
[Crossref] [Google Scholar] [PubMed]
- Hubatsch I, Ragnarsson EG, Artursson P. Determination of drug permeability and prediction of drug absorption in Caco-2 monolayers. Nat Protoc 2007;2(9):2111-9.
[Crossref] [Google Scholar] [PubMed]
- Ghafourian T, Amin Z. QSAR models for the prediction of plasma protein binding. Bioimpacts 2013;3(1):21-7.
[Crossref] [Google Scholar] [PubMed]
- Daneman R, Prat A. The blood–brain barrier. Cold Spring Harb Perspect Biol 2015;7(1):a020412.
- Pecoraro B, Tutone M, Hoffman E, Hutter V, Almerico AM, Traynor M. Predicting skin permeability by means of computational approaches: Reliability and caveats in pharmaceutical studies. J Chem Informat Model 2019;59(5):1759-71.
[Crossref] [Google Scholar] [PubMed]